BEETHOVEN CLASSIC


Mechanistic insights into the specificity of glycosaminoglycan interactions with regulatory proteins
- Funding Programme: BEETHOVEN CLASSIC from National Science Centre (Poland) and German Research Council
- Duration: 31.08.2020-30.08.2023
- Budget: 976 500 PLN (232 500 EUR)
- Project Leader: Sergey Samsonov
- Link to project description on NCN (Narodowe Centrum Nauki)
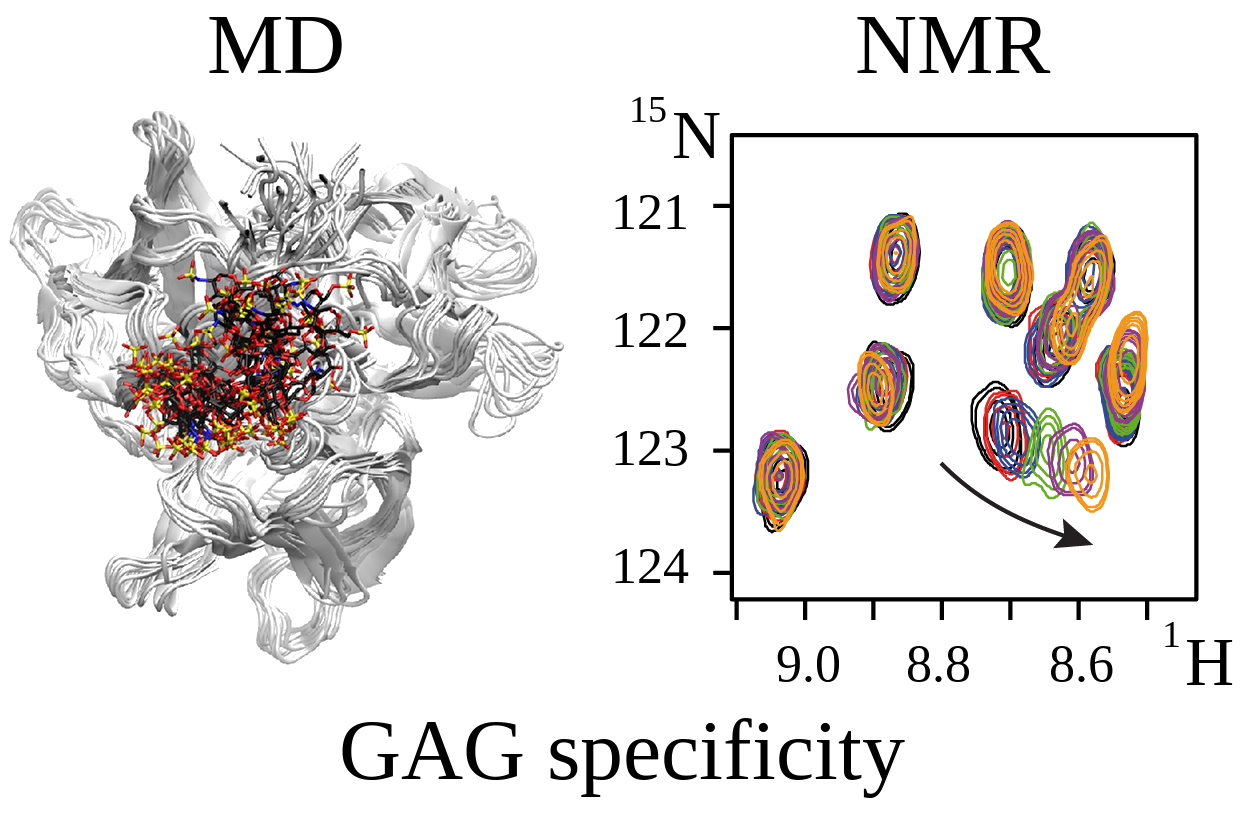
In the proposed project, we aim to answer the central question in the field of protein-glycosaminoglycan interactions by
establishing the specificity of the intermolecular recognition in these biologically relevant systems using
complementary molecular modeling and NMR approaches. The data obtained in this project will be useful for the fundamental
understanding of these systems,
which is a key for molecular-based rational design of new strategies in tissue regeneration therapies.
Glycosaminoglycans (GAGs) represent a class of linear anionic periodic polysaccharides made up of disaccharide
repetitive units containing a hexosamine and an uronic acid. These molecules present varying net degrees and patterns o
f sulfation. GAGs are major components of the extracellular matrix and play important roles in numerous cellular
processes such as signaling, anticoagulation, angiogenesis and communication. Their biological role is executed
though intermolecular interactions with protein partners such as growth factors, chemokines, proteases and collagen.
Disruptions of protein-GAG interactions can cause a variety of pathologies including cancer, Alzheimer’s and prion
diseases, autoimmune and inflammatory disorders. Many of these properties render GAGs promising molecules for the
design of new biomaterials in regenerative therapies. At the same time, protein-GAG interactions are still not well
characterized at the molecular level. Classical structure determination techniques as well as computational approaches
face challenges when dealing with these systems because of GAG’s particular properties as limited availability of
experimental structures, extensive length, flexibility, periodicity, symmetry, multipose binding and high variety in
the sulfation pattern. The latter is suggested to be crucial for defining their recognition by protein targets and
the involvement in particular biochemical processes. Among these challenges, there is one central question in the
protein-GAG research field that remains unanswered: How specific are protein-GAG interactions? For several systems,
it was shown that the alterations in the GAG type and sulfation pattern changes their protein binding capabilities
specifically, whereas for others, it is demonstrated that the only parameter that affects the strength of such
interactions is a GAG charge, and so these interactions are completely driven by electrostatics and are, therefore,
unspecific. However, there are no systematic studies dealing with the question of protein-GAG specificity.
In our project, we aim to get deeper insights into the specificity in protein-GAG interactions by:
1) characterization of the interactions between interleukine-8 (IL-8), a well-known regulatory protein,
with GAGs and other negatively charged ligands of different chemical origin (i.e., a series of anionic peptides,
DNA, ATPγS, anionic polymers) with different flexibility and charge density which could potentially mimic GAGs;
2) analysis of GAGs interactions with cathepsin B, a protein representing a family of proteases.
In spite of featuring the same fold, all members of the cathepsin family interact differently with GAGs in terms
of the corresponding complex structures, binding affinities and enzymatic activity impacts of these interactions.
We have carried out a solid amount of preliminary work relevant to the current proposal. In previous cooperative
studies performed by the applicants, 6 out of 8 common publications dealt with the analysis of IL-8/GAG systems.
Dr. Samsonov, in addition, participated in two publications devoted to cathepsins and their interactions with GAGs.
In the project, we will use the combination of molecular modeling at the University of Gdańsk and nuclear
magnetic resonance (NMR) at Leipzig University based approaches such as molecular docking, molecular dynamics,
free energy calculations and heteronuclear single quantum coherence (HSQC) spectroscopy titration, paramagnetic
resonance enhancement (PRE), pseudocontact chemical shift (PCS) methods, respectively.
The data obtained in this project will contribute to the fundamental understanding of the protein-GAG
biologically relevant systems providing mechanistic insights into molecular interactions within these
systems. In addition, the systematic application of the combined modeling and experimental NMR approaches to
analyze protein-GAG interactions will allow for the development and refinement of a general pipeline for analysis
of these systems. Altogether, the results planned within the project will be potentially useful for rational
establishing of novel approaches in the field of tissue regeneration which are based on fundamental knowledge of
protein-GAG systems at the atomistic level.
As we have already shown in our previous collaboration, a combination of molecular modeling with NMR-based
methods proved to be very beneficial and complementary allowing to obtain results that are not accessible
by either of the techniques alone. Whereas NMR could assist to validate and guide the calibration of computational
approaches, modeling provides atomistic details and mechanistic explanation of the experimental data.
The use of modeling and experimental approaches together, therefore, reduces the time and financial support
needed for reaching the research objectives. Last but not least, such a bilateral cooperation would contribute
to the scientific development of the Ph.D. students involved in the project by providing them a brilliant
opportunity to participate in an interdisciplinary international research project and so to broaden their
scientific expertise.
